MISST - Microstructure Imaging Sequence Simulation ToolBox
Introduction
Microstructure Imaging Sequence Simulation Toolbox (MISST) is a practical diffusion MRI simulator for development, testing, and optimisation of novel MR pulse sequences for microstructure imaging. MISST is based on a matrix method approach and simulates the signal for a large variety of pulse sequences and tissue models. Its key purpose is to provide a deep understanding of the restricted diffusion MRI signal for a wide range of realistic, fully flexible scanner acquisition protocols, in practical computational time.
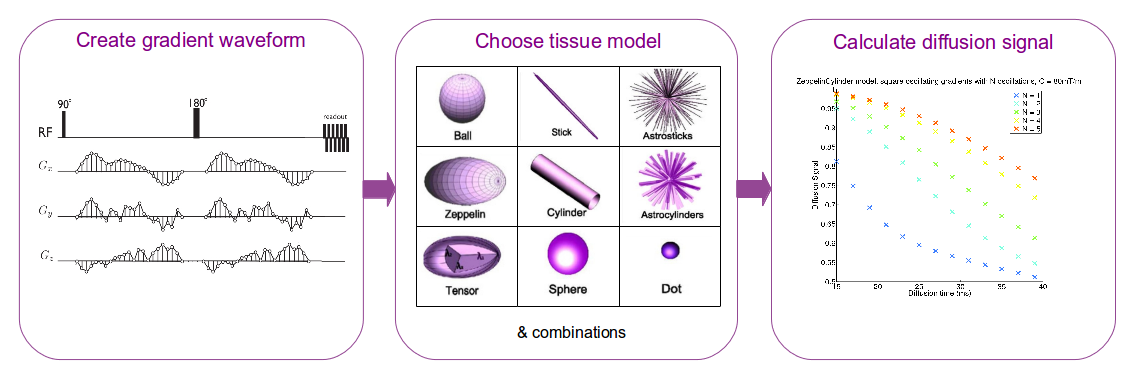
The novelty of MISST is that it simulates diffusion MRI signal with any user-defined diffusion gradient sequence from a standard PGSE to more advanced sequences such as oscillating gradients, dPFG, q-mas, STEAM, etc. The diffusion signal is calculated using the matrix formalism ( Callaghan 1997 ) and the implementation detailed in Drobnjak et al. 2010 and Drobnjak et al. 2011. The existing parametrizations for square and trapezoidal oscillating gradients follow the description in Ianus et al. 2012. The diffusion substrates available in MISST are the basic compartments presented in Panagiotaki et al. 2012, as well as some two- and three-compartment models of white matter.
Target audience
MISST is designed for diffusion MRI researchers who are interested in understanding and developing diffusion pulse sequences for imaging microstructure.
Download
Terms and Conditions
Step-by-step getting started tutorial
- Include MISST toolbox in the MATLAB search path:
/usr/local/MISST, then run
addpath(genpath('/usr/local/MISST'))
- Example 1: square oscillating gradients
Create an example protocol with square oscillating gradients
wave_form function. The available sequences are:
% add some combinations of parameters:
clear all
delta = 0.015:0.005:0.04; % duration in s
smalldel = delta - 0.005; % duration in s
Nosc = 1:5; % number of lobes in the oscillating gradient. A full period has 2 lobes
G = 0.08; % gradient strength in T/m;
tau = 1E-4; % time interval for waveform discretization
it = 0;
protocol_init.pulseseq = 'SWOGSE';
for i = 1:length(delta)
for j = 1:length(Nosc)
it = it+1;
protocol_init.smalldel(it) = smalldel(i);
protocol_init.delta(it) = delta(i);
protocol_init.omega(it) = Nosc(j).*pi./smalldel(i);
protocol_init.G(it) = G;
protocol_init.grad_dirs(it,:) = [1 0 0]; % gradient in x direction
end
end
protocol_init.tau = tau;
protocolGEN.pulseseq = 'GEN'; protocolGEN.G = wave_form(protocol_init); protocolGEN.tau = tau; % include smalldel and delta as they make computation slightly faster protocolGEN.delta = protocol_init.delta; protocolGEN.smalldel = protocol_init.smalldel;
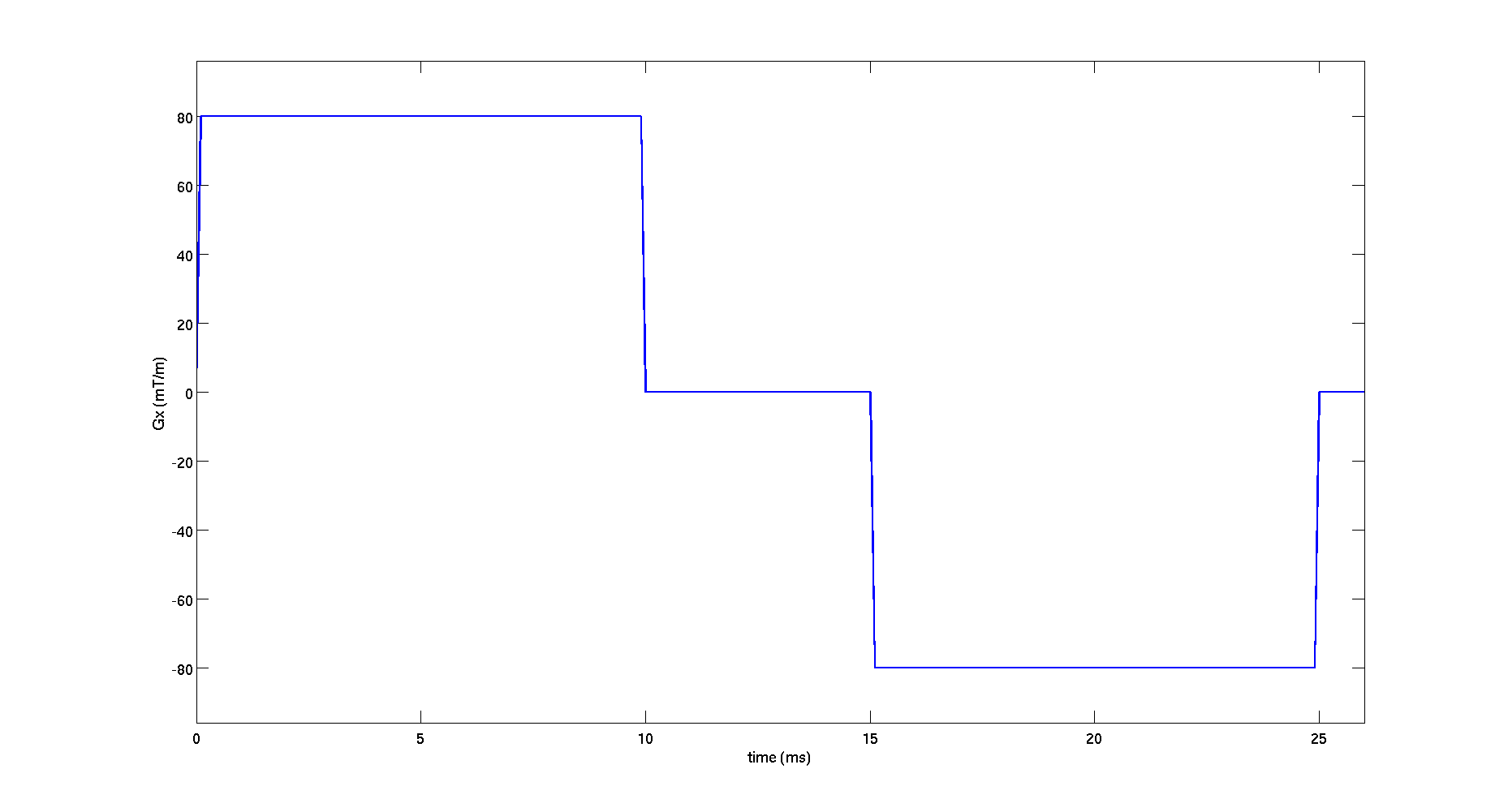
- View the first waveform in the protocol
figure();
Gx = protocolGEN.G(1,1:3:end);
plot((0:tau:(length(Gx)-1)*tau)*1E3,Gx*1000,'LineWidth',2)
xlim([0,(protocolGEN.smalldel(1)+protocolGEN.delta(1)+10*tau)*1E3])
ylim([min(Gx)*1200 max(Gx)*1200])
xlabel('time (ms)','FontSize',16);
ylabel('Gx (mT/m)','FontSize',16);
set(gca,'FontSize',16);
- Synthesize the diffusion signal for this protocol
model.name = 'ZeppelinCylinder'; di = 1.7E-9; % intrinsic diffusivity dh = 1.2E-9; % hindered diffusivity rad = 4E-6; % cylinder radius % angles in spherical coordinates describing the cylinder orientation; theta = 0; % angle from z axis phi = 0; % azimuthal angle ficvf = 0.7; % intracellular volume fraction model.params = [ficvf di dh rad theta phi];
protocolGEN = MMConstants(model,protocolGEN);
signal = SynthMeas(model,protocolGEN);
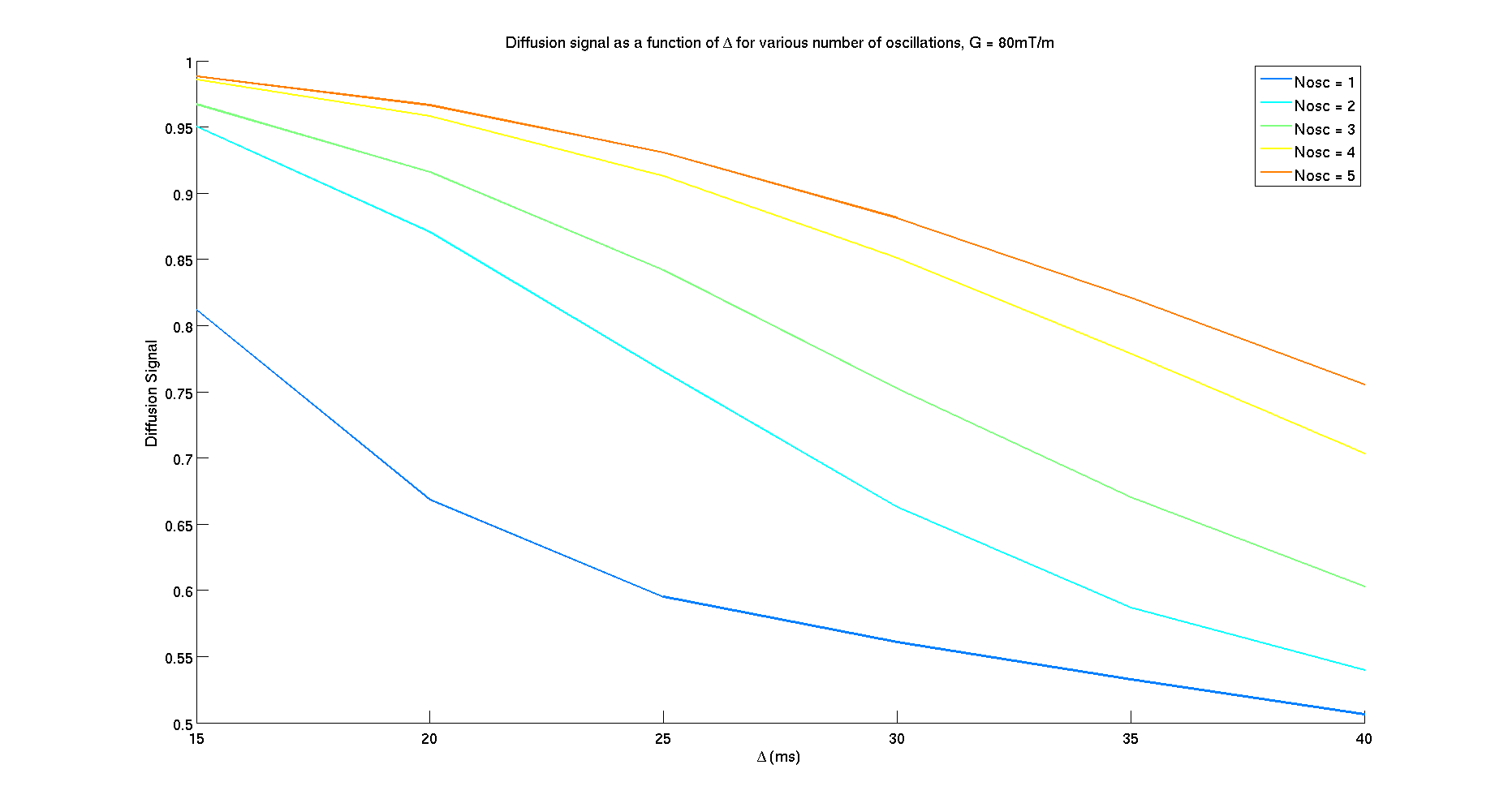
- Plot the synthesized diffusion signal
signal_matrix = reshape(signal,length(Nosc),length(delta));
figure();
colors = jet(length(Nosc));
hold on;
for i = 1:length(Nosc)
legend_name{i} = ['Nosc = ' num2str(Nosc(i))];
plot(delta*1000,signal_matrix(i,:),'LineWidth',2,'Color',colors(i,:));
end
xlabel('\Delta (ms)','FontSize',16);
ylabel('Diffusion Signal','FontSize',16);
set(gca,'FontSize',16);
title('Diffusion signal as a function of \Delta for various number of oscillations, G = 80mT/m')
legend(legend_name,'FontSize',16)
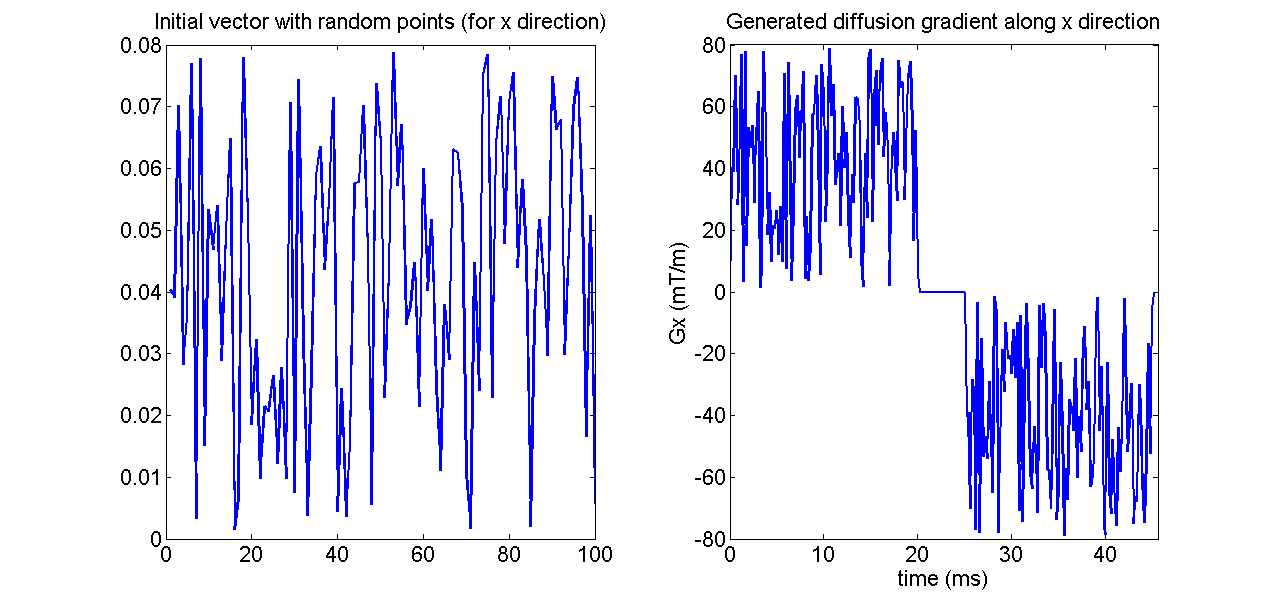
- Example 2: random gradient waveform. To show the true potential of MISST which can work with any diffusion sequence, for this example we chose a diffusion gradient given by 3 vectors of random numbers for gradients in x,y and z directions.
clear all smalldel = 0.02; % duration of the waveform; Npoints = 100; % 100 random points tau = smalldel / Npoints; % sampling interval p180 = 0.005; % time interval between the first and second waveforms (duration of rf pulse) Gmax = 0.08; % maximum gradient in T/m; % the gradient waveform will be repeated after the 180 pulse wf = [rand(1,Npoints)*Gmax; rand(1,Npoints)*Gmax; rand(1,Npoints)*Gmax]; % gradient waveform consists of random points along x, y and z % create protocolGEN in the necessary format protocolGEN.G = [zeros(1,3) wf(:)' zeros(1,3* round(p180/tau)) -wf(:)' zeros(1,3)]; protocolGEN.tau=tau; protocolGEN.pulseseq = 'GEN';
figure();
subplot(1,2,1)
plot(wf(1,:),'LineWidth',2)
title('Initial vector with random points (for x direction)','FontSize',16);
set(gca,'FontSize',16);
subplot(1,2,2)
Gx = protocolGEN.G(1,1:3:end);
plot((0:protocolGEN.tau:(length(Gx)-1)*protocolGEN.tau)*1E3,Gx*1000,'LineWidth',2)
xlabel('time (ms)','FontSize',16);
ylabel('Gx (mT/m)','FontSize',16);
title('Generated diffusion gradient along x direction','FontSize',16);
set(gca,'FontSize',16);
xlim([0,(length(Gx)+1)*tau*1E3])
model.name = 'ZeppelinCylinder'; di = 1.7E-9; % intrinsic diffusivity dh = 1.2E-9; % hindered diffusivity rad = 4E-6; % cylinder radius % angles in spherical coordinates describing the cylinder orientation; theta = 0; % angle from z axis phi = 0; % azimuthal angle ficvf = 0.7; % intracellular volume fraction model.params = [ficvf di dh rad theta phi];
protocolGEN = MMConstants(model,protocolGEN);
signal = SynthMeas(model,protocolGEN);
- Example 3: diffusion sequences used in Drobnjak et al. 2011 .
1. dPFG sequences for parameters used in Figure 1b from Drobnjak et al. 2011.
clear all
Npoints = 25;
protocoldPFG1.pulseseq = 'dPFG';
protocoldPFG1.smalldel = [repmat(0.0015,1,Npoints) repmat(0.0045,1,Npoints) ...
repmat(0.0075,1,Npoints)];
protocoldPFG1.G = [repmat(0.3757,1,Npoints) repmat(0.1252,1,Npoints) ...
repmat(0.0751,1,Npoints)];
protocoldPFG1.delta = repmat(0.04,1,Npoints*3);
protocoldPFG1.theta = repmat(pi/2,1,Npoints*3);
protocoldPFG1.phi = repmat(0,1,Npoints*3); % first gradient along x direction
protocoldPFG1.theta1 = repmat(pi/2,1,Npoints*3);
% second gradient rotating in x-y plane
protocoldPFG1.phi1 = repmat(linspace(0,2*pi,Npoints),1,3);
protocoldPFG1.tm = repmat(0,1,Npoints*3);
tau = 1E-4;
protocoldPFG1.tau = tau;
protocolGEN.pulseseq = 'GEN'; protocolGEN.G = wave_form(protocoldPFG1); protocolGEN.tau = tau;
figure();
G_plot = protocolGEN.G(1,1:3:end);
plot((0:tau:(length(G_plot)-1)*tau)*1E3,G_plot*1000,'LineWidth',2)
xlim([0,(length(G_plot)+5)*tau*1E3])
ylim([min(G_plot)*1200 max(G_plot)*1200])
set(gca,'FontSize',16);
xlabel('time (ms)','FontSize',16);
ylabel('Gx (mT/m)','FontSize',16);
title('Gradient wavefor for \phi = 0')
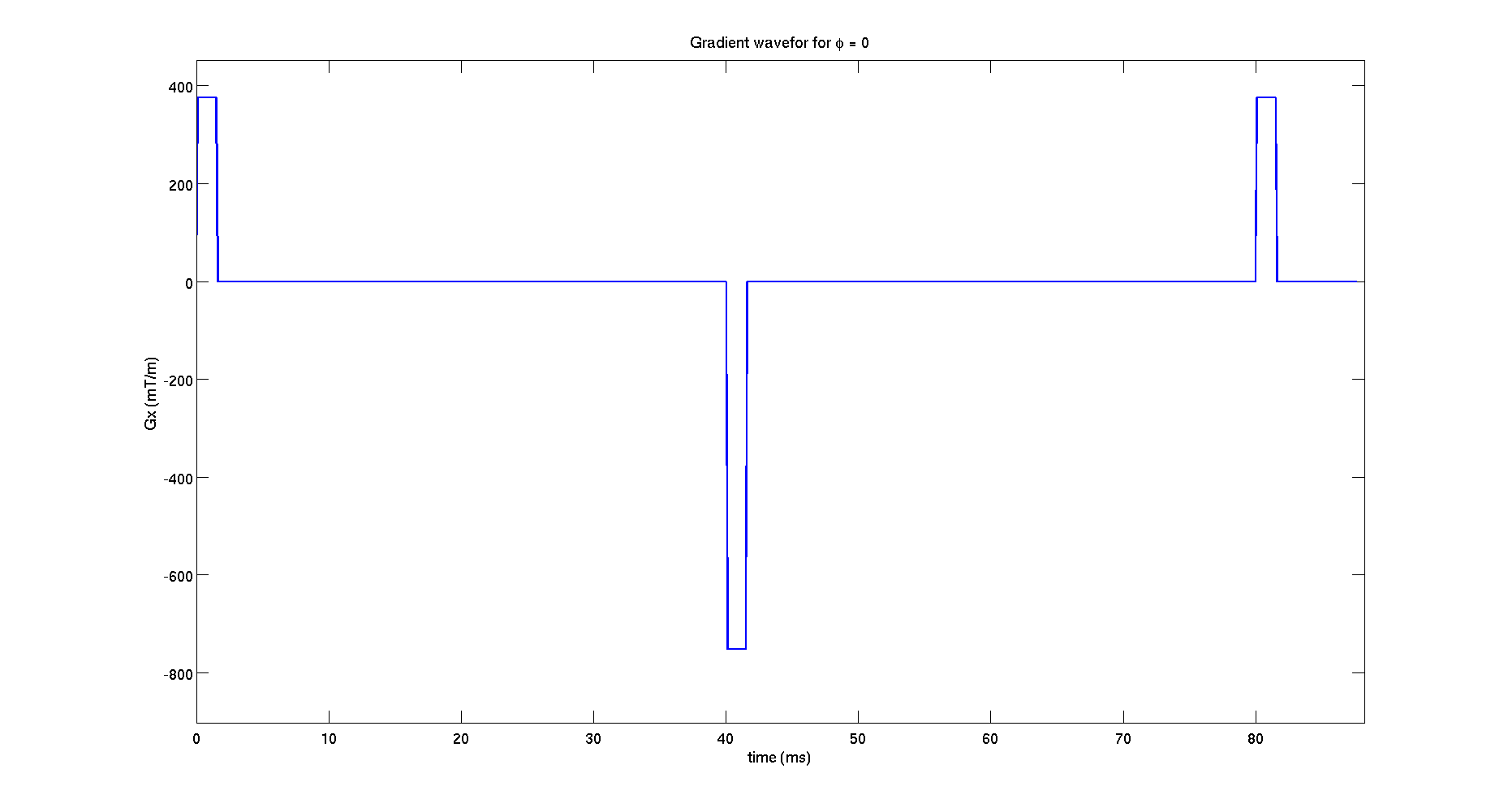
model.name = 'Cylinder'; di = 2E-9; % intrinsic diffusivity rad = 5.2E-6; % cylinder radius % angles in spherical coordinates describing the cylinder orientation; theta = 0; % angle from z axis phi = 0; % azimuthal angle model.params = [ di rad theta phi];
protocolGEN = MMConstants(model,protocolGEN);
signal = SynthMeas(model,protocolGEN);
signal_plot = reshape(signal,Npoints,3);
figure();
hold on;
plot(linspace(0,2*pi,Npoints),signal_plot(:,1),'xr',...
linspace(0,2*pi,Npoints),signal_plot(:,2),'sb',...
linspace(0,2*pi,Npoints),signal_plot(:,3),'<g',...
'LineWidth',2,'MarkerSize',12);
xlabel('\phi','FontSize',16);
ylabel('Diffusion Signal','FontSize',16);
legend('\delta = 1.5 ms','\delta = 4.5 ms','\delta = 7.5 ms')
set(gca,'FontSize',16);
title('Diffusion signal for dPFG (Fig. 1b)')
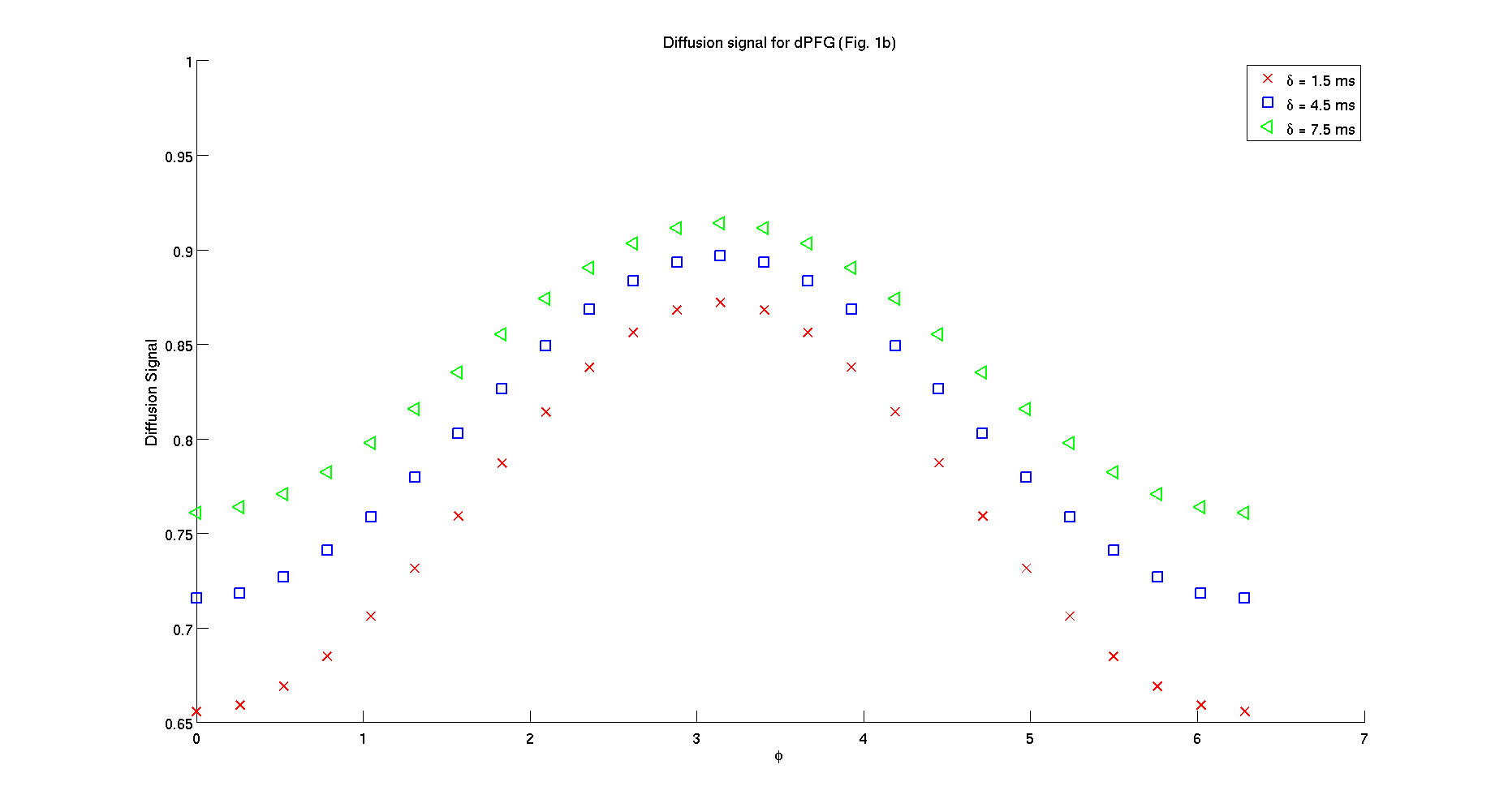
- 2. Helical waveforms for parameters used in Figure 2b from Drobnjak et al. 2011.
clear all
fosc = 0.2:0.2:3; % normalized frequency
smalldel = 0.01; % duration
delta = 0.0127;
protocolHelical.pulseseq = 'Helical';
protocolHelical.smalldel = repmat(smalldel,1,length(fosc)*3) ;
protocolHelical.G = [repmat(0.05,1,length(fosc)) repmat(0.08,1,length(fosc))...
repmat(0.15,1,length(fosc))];
protocolHelical.delta = repmat(delta,1,length(fosc)*3);
protocolHelical.omega = repmat(2*pi*fosc/smalldel,1,3);
protocolHelical.slopez = repmat(1/5/smalldel,1,length(fosc)*3);
tau = 1E-4;
protocolHelical.tau = tau;
protocolGEN.pulseseq = 'GEN'; protocolGEN.G = wave_form(protocolHelical); protocolGEN.tau = tau;
figure();
G_plot = protocolGEN.G(end,1:3:end);
plot((0:tau:(length(G_plot)-1)*tau)*1E3,G_plot*1000,'LineWidth',2)
xlim([0,(length(G_plot)+5)*tau*1E3])
ylim([min(G_plot)*1200 max(G_plot)*1200])
xlabel('time (ms)','FontSize',16);
ylabel('Gx (mT/m)','FontSize',16);
set(gca,'FontSize',16);
title('Gradient waveform in x direction with the highest frequency','FontSize',16);
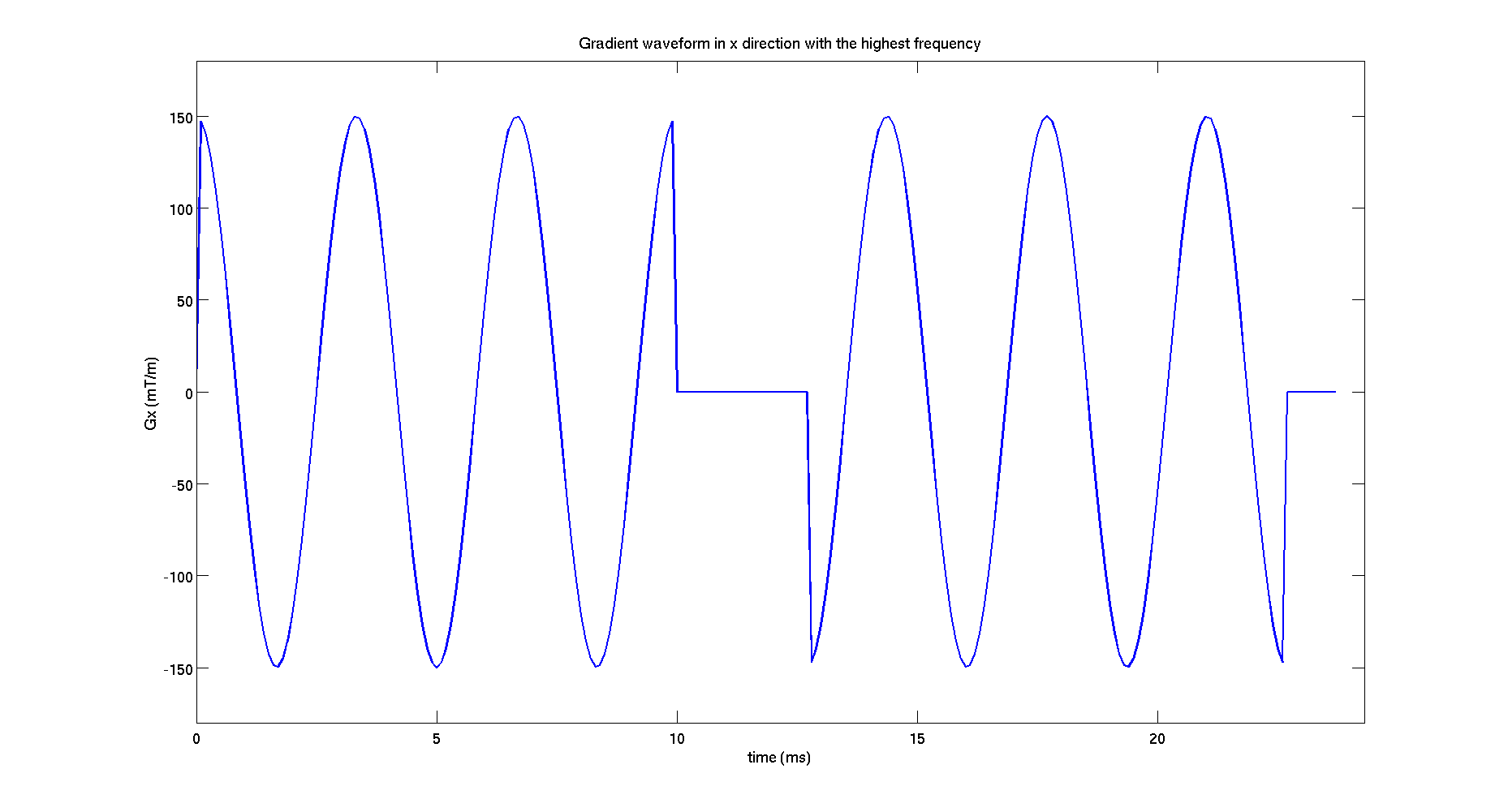
model.name = 'Cylinder'; di = 2E-9; % intrinsic diffusivity rad = 5E-6; % cylinder radius % angles in spherical coordinates describing the cylinder orientation; theta = 0; % angle from z axis phi = 0; % azimuthal angle model.params = [ di rad theta phi];
protocolGEN = MMConstants(model,protocolGEN);
signal = SynthMeas(model,protocolGEN);
signal_plot = reshape(signal,length(fosc),3);
figure();
hold on;
plot(fosc,signal_plot(:,1),'xr',...
fosc,signal_plot(:,2),'sb',...
fosc,signal_plot(:,3),'<g',...
'LineWidth',2,'MarkerSize',12);
xlabel('f','FontSize',16);
ylabel('Diffusion Signal','FontSize',16);
legend('G = 50 mT/m','G = 80 mT/m','G = 150 mT/m')
set(gca,'FontSize',16);
title('Diffusion signal for Helical waveform (Fig. 2b)')
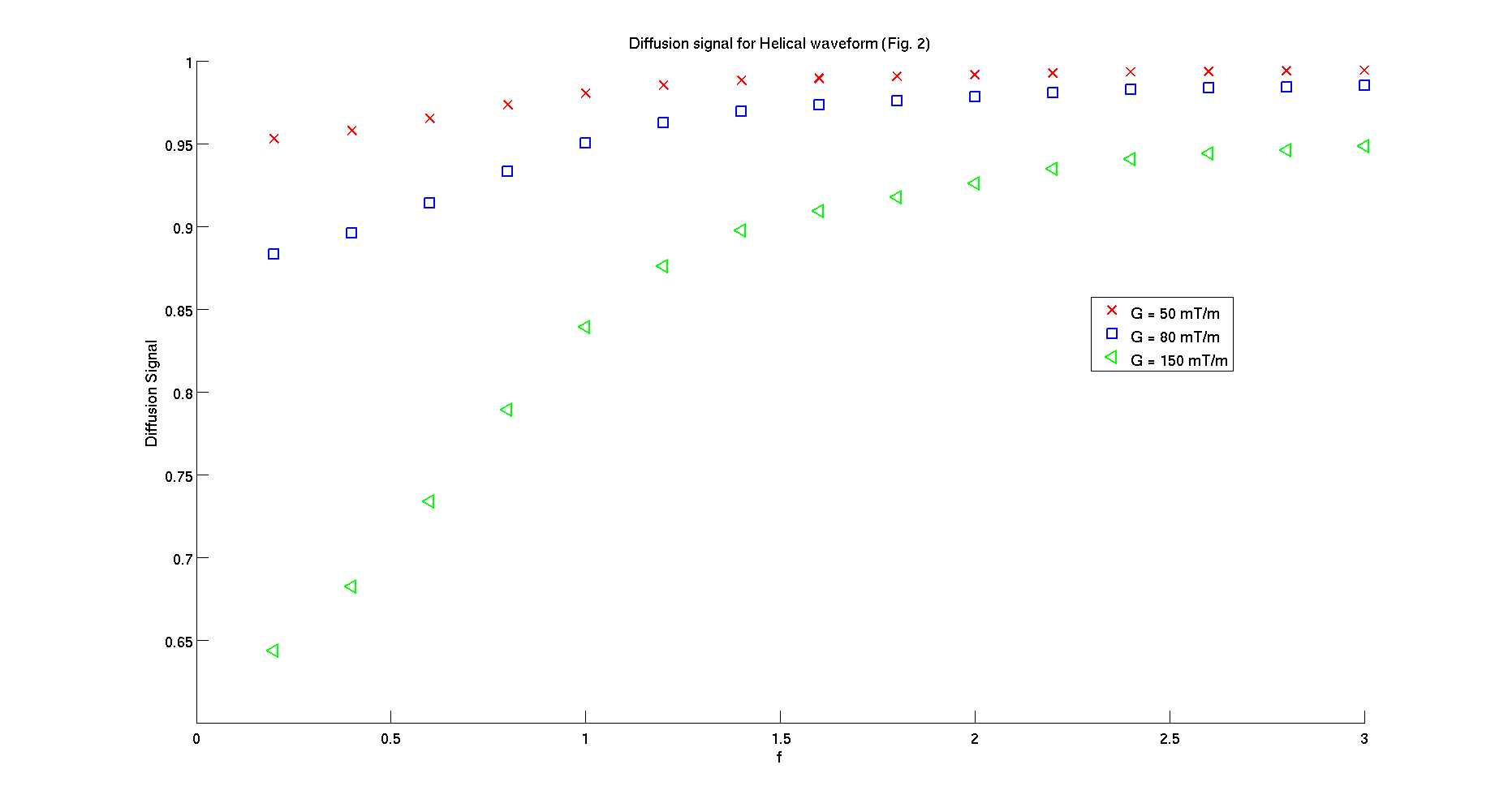
- 3. STEAM sequences for parameters used in Figure 3b from Drobnjak et al. 2011.
clear all
protocolSTEAM.pulseseq = 'STEAM';
protocolSTEAM.smalldel = repmat(0.0079,1,50);
protocolSTEAM.sdels = repmat(0.001,1,50);
protocolSTEAM.sdelc = repmat(0.0015,1,50);
protocolSTEAM.G = repmat(0.14,1,50);
protocolSTEAM.Gs = repmat(0.139,1,50);
protocolSTEAM.Gc = repmat(0.04,1,50);
protocolSTEAM.grad_dirs = abs(ReadCaminoElecPS(sprintf('PointSets/Elec%03i.txt',50)));
protocolSTEAM.tau1 = repmat(0,1,50);
protocolSTEAM.tau2 = repmat(0,1,50);
protocolSTEAM.tm = repmat(0.01,1,50);
tau = 1E-4;
protocolSTEAM.tau = tau;
protocolGEN.pulseseq = 'GEN'; protocolGEN.G = wave_form(protocolSTEAM); protocolGEN.tau = tau;
figure();
Gz = protocolGEN.G(1,3:3:end);
plot((0:tau:(length(Gz)-1)*tau)*1E3,Gz*1000,'LineWidth',2)
xlim([0,(length(Gz)+5)*tau*1E3])
ylim([min(Gz)*1200 max(Gz)*1200])
xlabel('time (ms)','FontSize',16);
ylabel('Gz (mT/m)','FontSize',16);
set(gca,'FontSize',16);
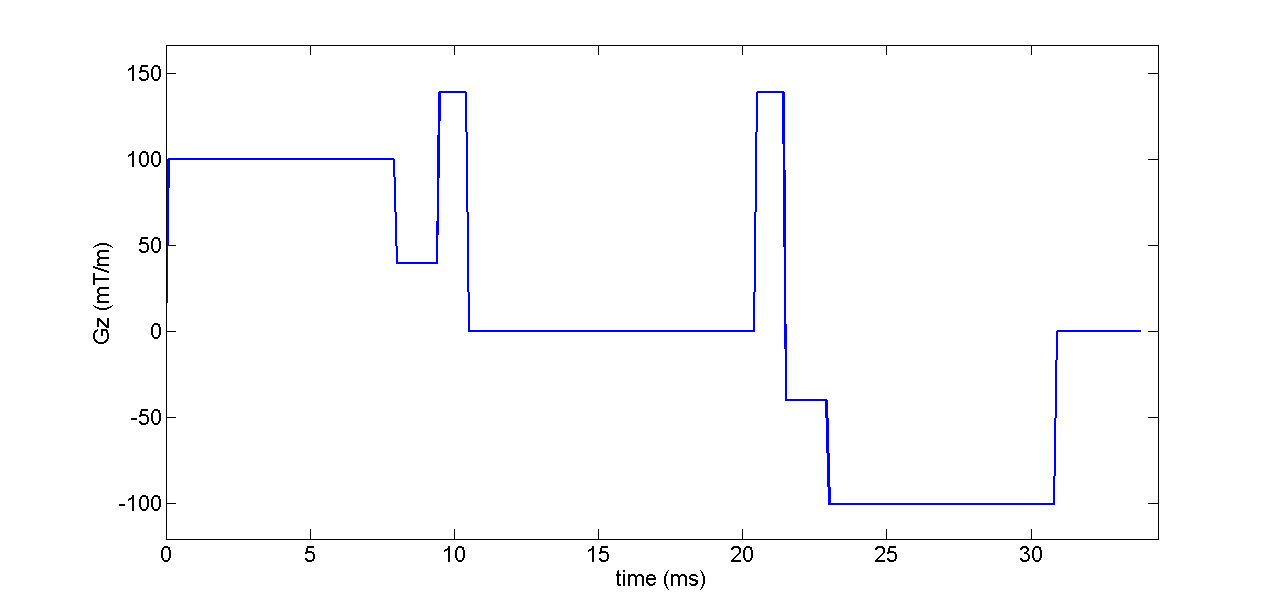
model.name = 'Cylinder'; di = 0.7E-9; % intrinsic diffusivity rad = 5E-6; % cylinder radius % angles in spherical coordinates describing the cylinder orientation; theta = 0; % angle from z axis phi = 0; % azimuthal angle model.params = [ di rad theta phi];
protocolGEN = MMConstants(model,protocolGEN);
signal = SynthMeas(model,protocolGEN);
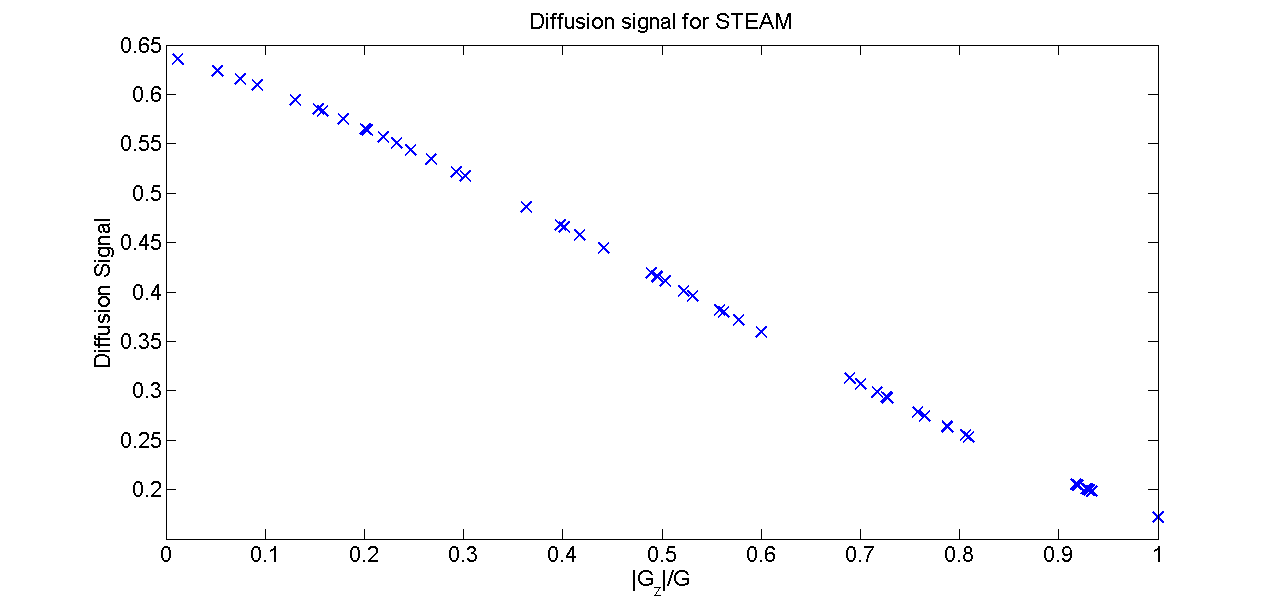
signal_plot = signal;
Gplot = abs(protocolSTEAM.grad_dirs(:,3));
figure();
plot(Gplot,signal_plot,'x','LineWidth',2,'MarkerSize',12);
xlabel('|G_z|/G','FontSize',16);
ylabel('Diffusion Signal','FontSize',16);
set(gca,'FontSize',16);
title('Diffusion signal for STEAM Fig. 3b')
- To help you start, the same commands are part of the scripts in the folder MISST/example
MISST tutorials
- Understand the format of the gradient waveform necessary for the simulation:
protocol.pulseseq = 'GEN' - the name of the sequence, -->protocol.G - discrete gradient waveforms which contain the gradient components (Gx , Gy and Gz ) at each point in time from the beginning of the measurement until the readout
protocol.tau - time interval between two points on the gradient waveform
protocol.smalldel - gradient duration
protocol.delta - time interval between the onset of the first and second pulse.
protocol.mirror = 0 (default value) or reflected protocol.mirror = 1 .
protocol.G is a M x 3K matrix which stores the values of the diffusion gradient at each time point. Each row represents one measurement and contains the values of the gradients in x, y and z direction. M is the total number of measurements and K is the number of gradient points in each direction for one measurement:
protocol.G includes the complete diffusion gradient and the user should take into account any realistic situations such as the duration of the rf pulses, crusher gradients, etc.
protocol.G should satisfy the echo condition, that the integral of the gradient over time should be 0.
- Understand the tissue models available in the simulation:
All the information related to the diffusion substrate is stored in a MATLAB structure, which we call “model” in the examples. For the purpose of signal generation, “model” has only two fields: model.name – the name of the model (listed above) and model.params - the values of the model parameters in S.I. units (m, s, etc.) of each model. See documentation for details.

