Our research on microstructure imaging has the ultimate goal of non-invasive histology. Classical histology uses a microscope to visualize the cellular architecture of tissue from a thinly sliced sample mounted on a slide. Histology provides the gold-standard diagnosis for a wide range of diseases from cancers to dementias. However, it requires a tissue sample from a biopsy or taken post mortem. The idea of non-invasive histology is to obtain the same information that a histologist gets from microscope images from non-invasive imaging techniques such as magnetic resonance imaging (MRI). The non-invasive nature of microstructure imaging avoids patient discomfort and possible side effects of standard biopsy-histology procedures and enables temporal monitoring through repeat measurements. Moreover, microstructure imaging produces maps over whole organs rather than a targeted biopsy sample of a few mm3, so is less prone to false negatives from poor targeting, and provides a more complete picture of heterogeneous diseases, such as cancer.
How do we do it?
In a nutshell
Microstructure imaging works by fitting mathematical models of cellular architecture to imaging data. Most of our work uses diffusion MRI, but other kinds of measurement can contribute. We solve an inverse problem in each image voxel to estimate and map histological tissue parameters; the figure below shows an example from the ActiveAx technique, which maps features of white matter such as axon diameter and density.
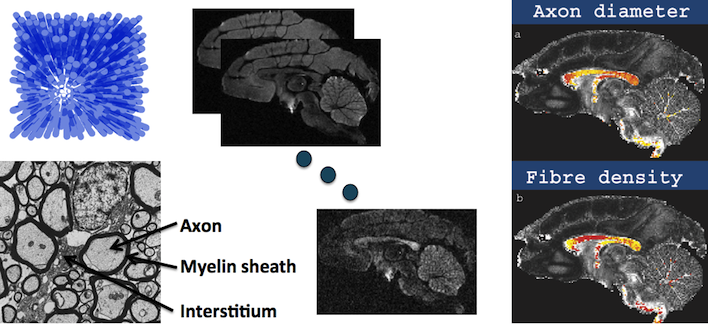
ActiveAx: microstructure imaging of white matter. Left shows a histology image of white matter (bottom), which is a cross-sectional view through a bundle of axon fibres, and a simple geometric model (top) consisting of parallel cylindrical axons. We acquire various diffusion images (middle) and fit the model parameters (cylinder size and density) in each voxel to obtain overlays of axon diameter (right top) and density (right bottom); red is low, yellow is high.
Active Imaging
Our approach centres on the computational modelling paradigm we call Active Imaging. The four key components are:

- Modelling: constructing computational models of how different features of the tissue microstructure affect the measurements we get from imaging devices.

- Sampling: deciding which set of measurements from which imaging devices provide the most information about the tissue microstructure given constraints on imaging time or cost.

- Fitting: estimating model parameters from sets of measurements.

- Validation: confirming that the estimates we make reflect the true state of the tissue.
Imaging techniques
We have particular expertise in an imaging technique called diffusion MRI. Diffusion MRI provides a unique probe into the microstructure of materials. The technique sensitizes the MR image intensity to the dispersion pattern of particles, usually water molecules, within tissue. Tissue microstructure, such as cell walls and membranes, impede the mobility of water and determine the dispersion pattern. Thus from multiple MR measurements sensitized to different aspects of the dispersion pattern, we can infer features of the tissue microstructure like cell size, shape and packing density. Although diffusion MRI is a lynchpin technique, many other non-invasive imaging techniques also contribute: other MRI modalities, such as relaxometry, elastography, and quantitative magnetization transfer; computed tomography (CT); positron emission tomography (PET); and optical imaging.
Where are we?
In a nutshell
We have developed a range of “multiple-fibre reconstruction” or “high angular resolution diffusion imaging (HARDI)” techniques for estimating fibre orientations and configurations from diffusion MRI enabling non-invasive studies of brain connectivity through tractography. Most of that work was with the support of the EPSRC Multiple Fibre Reconstructions project.
We have two primary working microstructure imaging techniques: ActiveAx and NODDI (Neurite Orientation Dispersion and Density Imaging), both designed for brain imaging. ActiveAx uniquely maps orientationally invariant axon diameter and density indices, as shown in the figure at the top of this page. It works very well on fixed brain samples that we can image for a long time with powerful scanners and is rapidly gaining popularity for these applications. Although, we have demonstrated ActiveAx on live human volunteers, the imaging time is close to one hour and the axon-diameter contrast remains weak. We aim to gain clinical applicability in the near future by exploiting faster acquisition to reduce scan time and specialized diffusion-weighted pulse sequences to increase sensitivity. NODDI produces separate maps of dispersion of fibre orientations (how much fanning/bending/etc), fibre density, and partial volume with free water (e.g. cerebro-spinal fluid). The traditional fractional anisotropy (FA) index from diffusion tensor imaging (DTI) confounds these three effects; NODDI allows separate analysis of each. The technique also provides useful information in grey matter, such as the dispersion and density of the dendritic tree. NODDI requires only 10-30 mins acquisition time with standard pulse sequences so is clinically feasible. Both techniques were developed within the EPSRC Direct Measurements of Microstructure from MRI and EU CONNECT projects and open-source post-processing code is available for both.
A large body of work in microstructure imaging focuses on white matter in the brain. White matter is the cabling that supports communication throughout the brain. It consists of bundles of axon fibres. Axon fibres, which are a few microns in diameter, are much smaller than single voxels (3D pixels) of MR images, which are usually one or two millimetres across. Lots of work over the last 20-30 years aims to recover the orientations of white matter fibres for the purposes of reconstructing brain connectivity; see for example our work on multiple fibre reconstruction algorithms summarized in (Seunarine 2009). Early in-vitro work, e.g. (Stanisz 1998; Assaf 2008), shows the potential for microstructure imaging with diffusion MRI to recover histological features of nervous tissue, such as axon diameter and density. Three key limitations prevent direct translation of those techniques to the living human brain: i) they use scanners orders of magnitude more powerful than current human systems; ii) they use very long imaging times; iii) they assume knowledge of the fibre orientation. Our work (Alexander 2008) uses experiment design optimization to demonstrate in theory that we can overcome all three limitations. These ideas underpin the ActiveAx technique (Alexander 2010), which provides the first maps of axon diameter and density indices over the brains of live human volunteers. We have subsequently made significant advances in each stage of the Active Imaging paradigm:
Modelling: (Panagiotaki 2012) provides a taxonomy of mathematical models of diffusion in white matter, including models from the literature and many more, and evaluates their ability to explain rich MRI data sets from fixed tissue. The work verifies the choice of model in ActiveAx and (Ferizi 2012) further verifies the choice by repeating the experiment using in-vivo data. (Zhang 2011) demonstrates the influence of fibre orientation dispersion in axon diameter estimation and provides new models to accommodate the effect. The original ActiveAx provides meaningful parameter estimates only in major white matter tracts where fibres are straight and parallel; the new models extend the method to a much wider portion of white matter where fanning and banding are significant. The NODDI technique (Zhang 2012) simplifies the ActiveAx technique by dropping the axon diameter parameter to estimate fibre density and orientation dispersion. Whereas data for ActiveAx take about 1h to acquire, NODDI works very well with data acquired in just 15 mins so has much greater clinical viability. Moreover, the technique provides useful information in grey matter as well as white matter. In grey matter, the dispersion of dendrite orientations provides contrast between functional areas that translates into contrast from the diffusion MRI signal (Nagy 2012). (Prevost 2011) provides an example of the Active Imaging paradigm in other imaging modalities, in particular dynamic contrast enhanced CT. (Proverbio 2012) presents a joint model of diffusion NMR and optical imaging to improve pore size estimation.
Sampling: (Dyrby 2012) demonstrate and quantify the major benefits of increased gradient strength for microstructure imaging using ActiveAx of fixed brains. (Schneider 2010) show that the acquisition time for ActiveAx reduces to one half if the fibre orientation is known making the technique clinically viable in, for example, the corpus callosum or cortico-spinal tracts. (Alexander 2012) demonstrates benefits of stimulated echo (STEAM) diffusion MRI over spin echo in estimating diameters of large axons. The most significant advances in ActiveAx and similar techniques will come from moving away from the standard pulsed-gradient spin-echo (PGSE) diffusion MR pulse sequence. (Drobnjak 2010) search the space of all possible gradient waveforms for those that maximize sensitivity to pore size. Square wave oscillating gradients consistently emerge and theoretical results suggest orders of magnitude greater sensitivity from these waveforms than standard PGSE. Experimental work verifies the benefit in glass-capillary phantoms (Siow 2012a) and fixed brain tissue (Siow 2012b). Follow-up theoretical work (Drobnjak 2011a) extends the matrix formalism (Callaghan 1997) that underpins the optimization to 3D. That enables (Drobnjak 2011b) to provide compelling evidence that no benefit to the estimation of circular pore-size comes from varying the orientation of waveforms within individual pulse sequences. In particular, double wavevector sequences (Cory 1990) provide no additional sensitivity. (Ianus 2012) derives analytical approximations to the signal from square-wave oscillating gradient spin echo (SWOGSE), which compute the signal in a fraction of the time required for the matrix method, making SWOGSE ActiveAx a viable future prospect. Ivana Drobnjak’s Leverhulme fellowship project takes advanced sampling techniques, such as SWOGSE, from proof of principle to human in-vivo applications. This next generation of ActiveAx has great prospects for clinically viable axon diameter mapping.
Fitting: The key advance in fitting technology is to move away from voxel-by-voxel estimation of microstructure parameters to exploit spatial coherence: microstructure is usually similar in proximal voxels so we can pool data to improve estimates. (Morgan 2011) use a spline model of axon diameter variation across the corpus callosum to estimate the mid-sagittal trend less noisily that with voxel-by-voxel fitting. (Sherbondy 2010) show in simulation that by assuming consistency of microstructure along fibre tracts, we can improve microstructure parameter estimates, and resolve long-standing ambiguities in tractography, such as kissing versus crossing.
Validation: We have developed various tools for validation of microstructure imaging applications. (Hall 2009) presents a diffusion simulation system that supports arbitrary tissue environments, from packed cylinders with diameters reflecting white matter (Alexander 2010) to arbitrary mesh models derived from confocal or electron microscopy images (Panagiotaki 2010). (Siow 2012) construct white matter phantoms from glass capillaries; (Panagiotaki 2010) use asparagus as a biological phantom with similar pore structure to white matter; (Richardson 2012) construct a viable tissue chamber to allow imaging of excised tissue in in-vivo conditions for ten hours or more.
How is it useful?
In a nutshell
Target clinical applications include dementias, such as Alzheimer’s disease, multiple sclerosis, epilepsy, and a range of other neurological conditions. Many neurological diseases have hallmark histopathology such as neuronal loss, axonal degeneration, amyloid plaque deposition, and neurofibrillary tangles, which can affect image intensities in different ways potentially allowing microstructure imaging techniques to detect them and grade their severity. Another key application is solid cancers, such as prostate cancer, brain tumours, and breast cancer. Different grades of cancer have different cellular architecture, for example more malignant tumours often have higher cellularity (cell density) than benign. Again such features are amenable to the microstructure imaging approach, which we hope will lead to clinical diagnostic imaging techniques in the near future.
The MRI Biomarkers EPSRC Healthcare Partnerships project provides dedicated funding for a focused effort on developing microstructure imaging techniques for white matter diseases; specifically multiple sclerosis and spinal cord injury. This requires models of pathology in addition to the models of normal white matter we have developed so far. It represents the first step towards a new paradigm of disease specific microstructure imaging techniques. Within the CONNECT project, we have begun to develop models of the disruptions to grey matter that occur in prion diseases to try to explain and exploit observed signal changes in diffusion MRI. (Figini et al 2012) presents some early results.
Although designed initially for healthy tissue, NODDI is rapidly gaining popularity as a clinical tool. It is useful for improving tractography for neurosurgical planning, because it is able to recover fibre directions in areas of oedema where DTI can fail. The more specific indices of fibre dispersion and axon/dendrite density are being used for studies into dementia, brain and spinal cord injury, multiple sclerosis, and infant brain development.
We are starting also to extend these ideas to other types of tissue. Our work within the Intelligent Imaging program grant develops models for grading of cancer tumours by sensitivity of MRI to microstructural information usually obtained from biopsy and histology. Microstructural measurements from grey matter are useful for detecting anomalies in dementia and epilepsy patients. The cellular microstructure of muscle tissue provides useful information in diseases like dystonia and also for sports scientists. The broader microstructure imaging paradigm has many other potential applications both in biomedicine and beyond and we are keen to collaborate on new ideas!
References
- Alexander, D. C., Dyrby, T. B. (2012). Compensation for bias from unwanted gradient contributions in STEAM diffusion MRI. Proc. 20th International Society for Magnetic Resonance in Medicine. pp.1884
- Alexander, D. C., Hubbard, P. L., Hall, M. G., Moore, E. A., Ptito, M., Parker, G. J., Dyrby, T. B. (2010). Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage 52(4), 1374-1389.
- Alexander, D. C. (2008). A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magnetic Resonance in Medicine 60, 439-448
- Assaf, Y., Blumenfeld-Katzir, T., Yovel, Y., Basser, P.J. (2008) AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magnetic Resonance in Medicine. 59, 1347–1354.
- Callaghan, P. T. (1997) A simple matrix formalism for spin echo analysis of restricted diffusion under generalized gradient waveforms. Journal of Magnetic Resonance 129, 74–84.
- Cory, D. G.,Garroway, A.N., Miller, J.B. (1990) Applications of spin transport as a probe of local geometry. Polym. Prepr. 31 149–150.
- Drobnjak, I., Alexander, D. C. (2011) Optimising time-varying gradient orientation for microstructure sensitivity in diffusion-weighted MR. Journal of Magnetic Resonance 212(2), 344-354.
- Drobnjak, I., Zhang, H., Hall, M. G., Alexander, D. C. (2011). The matrix formalism for generalised gradients with time-varying orientation in diffusion NMR. Journal of Magnetic Resonance, 210(1), 151-157.
- Drobnjak, I., Siow, B., Alexander, D. (2010). Optimizing gradient waveforms for microstructure sensitivity in diffusion-weighted MR. Journal of Magnetic Resonance 206 (1), 41-51
- Dyrby, T. B., Baaré, W. F., Alexander, D. C., Jelsing, J., Garde, E., Søgaard, L. V. (2011). An Ex Vivo Imaging Pipeline for Producing High-Quality and High-Resolution Diffusion-Weighted Imaging Datasets. Human Brain Mapping 32(4), 544-563.
- Dyrby, T. B., Sogaard, L. V., Hall, M. G, Ptito, M., and Alexander, D. C. (2012). Contrast and stability of the axon diameter index from microstructure imaging with diffusion MRI. Magnetic Resonance in Medicine. In Press
- Hall, M. G., Alexander, D. C. (2009). Convergence and Parameter Choice for Monte-Carlo Simulations of Diffusion MRI. IEEE Transactions on Medical Imaging 28(9), 1354-1364
- Ferizi U, Panagiotaki E, Schneider T, Wheeler-Kingshott CAM, Alexander DC (2012) White Matter Models of In Vivo Diffusion MRI Human Brain Data: A Statistical Ranking. Proceeding of the 16th Conference on Medical Image Understanding and Analysis.
- Figini, M., Alexander, D. C., Fasano, F., Farina, L., Baselli, G., Tagliavini, F., Bizzi, A. (2012). Mathematical models of diffusion in prion disease. Italian chapter of the international society for magnetic resonance in medicine.
- Ianus, A., Alexander, D. C. (2012). Gaussian Phase Distribution Approximation of the Square Wave Oscillating Gradient Spin-Echo (SWOGSE) Diffusion Signal. Proc. 20th International Society for Magnetic Resonance in Medicine. pp.3561
- Morgan, G. L., Zhang, H., Whitcher, B., Alexander, D. C. (2011). A framework for modelling the regional variation of white matter microstructure. Proc 19th International Society for Magnetic Resonance in Medicine, pp. 1929.
- Nagy, Z., Alexander, D.C., Thomas, D.L., Weiskopf, N., Sereno, M. (2012) Discriminatng Cortcal Regions Using HARDI Data Proc. Human Brain Mapping.
- Panagiotaki, E., Schneider, T., Siow, B., Hall, M. G., Lythgoe, M. F., Alexander, D. C. (2012). Compartment models of the diffusion MR signal in brain white matter: A taxonomy and comparison. NeuroImage 59(3), 2241-2254
- Panagiotaki, E., Hall, M. G., Zhang, H., Siow, B., Lythgoe, M. F., Alexander, D. C. (2010). High-Fidelity Meshes from Tissue Samples for Diffusion MRI Simulations. Med Image Comput Comput Assist Interv (MICCAI), 404-411
- Prevost, R., Buckley, D. L., Alexander, D. C. (2010). Optimization of the DCE-CT protocol using active imaging. Proceedings of the 7th IEEE International Symposium On Biomedical Imaging (ISBI): From Nano to Macro
- Richardson, S., Siow, B., Batchelor, A. M., Lythgoe, M. F., and Alexander, D. C. (2012). A viable isolated tissue system: a tool for detailed MR measurements and controlled perturbation in physiologically stable tissue. Magnetic Resonance in Medicine. In Press. DOI: 10.1002/mrm.24410
- Schneider, T., Wheeler-Kingshott, C. A. M., Alexander, D. C. (2010). In-Vivo Estimates of Axonal Characteristics Using Optimized Diffusion MRI Protocols for Single Fibre Orientation. Med Image Comput Comput Assist Interv (MICCAI) 13(Pt 1), 183-190
- Seunarine, K. K., Alexander, D. C. (2009). Multiple fibers: beyond the diffusion tensor. In Diffusion MRI: from quantitative measurement to in vivo neuroanatomy ( pp.55-72). eds Johansen-Berg, H. and Behrens, T. E. J. Academic Press.
- Sherbondy, A. J., Rowe, M. C., Alexander, D. C. (2010). MicroTrack: an algorithm for concurrent projectome and microstructure estimation. Med Image Comput Comput Assist Interv (MICCAI)13(Pt 1), 183-190
- Siow, B., Drobnjak, I., Chatterjee, A., Lythgoe, M. F., Alexander, D. C. (2012). Estimation of pore size in a microstructure phantom using the optimised gradient waveform diffusion weighted NMR sequence. Journal of Magnetic Resonance 214, 51-60
- Siow, B., Ianus, A., Drobnjak, I., Lythgoe, M.F. and Alexander, D. C. (2012) Optimised oscillating gradient diffusion MRI for the estimation of axon radius in an ex-vivo rat brain. Proc. 20th International Society for Magnetic Resonance in Medicine. pp.357
- Stanisz, G.J., Szafer, A., Wright, G.A., Henkelman, M., 1997. An analytical model of restricted diffusion in bovine optic nerve. Magn. Reson. Med. 37, 103–111.
- Zhang, H., Schneider, T., Wheeler-Kingshott, C. A., Alexander, D. C. (2012). NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61(5), 1000-1016
- Zhang, H., Hubbard, P. L., Parker, G. J. M., Alexander, D. C. (2011). Axon diameter mapping in the presence of orientation dispersion with diffusion MRI. NeuroImage 56(3), 1301-1315

